http://www.jcancer.org/v10p1663.htm
Available studies demonstrate that receptor-type tyrosine-protein phosphatase zeta (PTPRZ1) is expressed in different tumor tissues, and functions in cell proliferation, cell adhesion and migration, epithelial-to-mesenchymal transition (EMT), cancer stem cells and treatment resistance by interacting with or binding to several molecules.
These included pleiotrophin (PTN), midkine, interleukin-34, β-catenin, VEGF, NF-κB, HIF-2, PSD-95, MAGI-3, contactin and ErbB4.
PTPRZ1 was involved in survival signaling and could predict the prognosis of several tumors. This review discusses: the current knowledge about PTPRZ1, its expression, co-receptors, ligands, functions, signaling pathway, prognostic values and therapeutic agents that target PTPRZ1.
Keywords: PTPRZ1, pleiotrophin, midkine, review
PTPRZ1 gene encodes receptor-type tyrosine-protein phosphatase zeta (PTPRZ1), which is expressed in different cancer tissues as either a chondroitin sulfate (CS) proteoglycan or a nonproteoglycan form [1, 2]. PTPRZ1 is overexpressed in several tumors, such as lung cancer, cervical cancer, hepatocarcinoma, renal cancer, and glioblastoma [3-7]. PTPRZ1 is localized not only in its normal association with the cell membrane but also scattered in cytoplasm and in nuclei in different breast cancer cells and, in the case of infiltrating ductal carcinomas, the distribution of PTPRZ1 changes as the breast cancer become more malignant[8]. PTPRZ1 has different functions in the cancer development, progression and metastasis, further influences the cancer treatment and prognosis. In order to comprehensively review the recent research on PTPRZ1 gene and PTPRZ1, we reviewed the relevant studies based on medical databases and analyzed its expression, prognostic values based on TCGA data.
Pubmed and EMBASE was comprehensively searched using “PTPZ” or “PTPRZ” or “RPTPB” or “PTP18” or “phosphacan” or “HPTPZ” or “RPTPbeta” or “HPTPzeta” or “R-PTP-zeta-2” or “PTP-ZETA” or “protein tyrosine phosphatase, receptor type Z1” or “RTP Type Z1” or “receptor-type tyrosine-protein phosphatase zeta” or “Protein-tyrosine phosphatase receptor type Z polypeptide 1” or “Protein-tyrosine phosphatase receptor type Z polypeptide 2”. There were no language, publication date, or publication types restrictions. The search was conducted at February 12, 2018, and updated at September 1, 2018. 345 citations from Pubmed and 356 citations from EMBASE were retrieved. TCGA (The Cancer Genome Atlas, https://cancergenome.nih.gov/) data was reviewed to analyze the expression of PTPRZ1 gene and PTPRZ1 gene mutations. The prognostic values of PTPTZ1 gene in different kinds of cancer were analyzed using GEPIA (http://gepia.cancer-pku.cn/) [9]. The protein-protein interaction (PPI) networks from available studies and public databases were conducted and visualized by the Cytoscape software.
Gene expression and mutations
The expression of PTPRZ1 gene, gene mutations, gene fusions were analyzed based on TCGA databases.Patients with glioblastoma multiforme (GBM) expressed the highest PTPRZ1 gene level, followed by brain lower grade glioma (LGG) and head and neck squamous cell carcinoma (Figure 1).
PTPRZ1 was up-regulated in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), GBM, LGG, lung squamous cell carcinoma (LUSC),
and down-regulated in breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), and stomach adenocarcinoma (STAD). The rates of PTPRZ1 gene mutations were different across tumors, and uterine corpus endometrial carcinoma (UCEC) (16.79%) has the highest rate of PTPRZ1 gene mutation, followed by lung adenocarcinoma (LUAD) (12.87%) and skin cutaneous melanoma (SKCM) (12.15%) (Figure 2).
Fusion genes combine parts of two or more original genes, generating from chromosomal rearrangement or abnormal transcription. About 25 PTPRZ1 fusion genes were found[10] (Table 1), but few of them were well studied.
PTPRZ1-MET fusion was found in 7.7% grade III astrocytomas (1/13) and 15.0% secondary glioblastomas (3/20), and tumors from patients harbouring PTPRZ1-MET-fused glioblastoma are resistant to temozolomide therapy and have significantly compromised overall survival rates [11, 12]. PTPRZ1-MET exosomes exhibit a more aggressive phenotype and induce epithelial-to-mesenchymal transition (EMT), enhance tumorigenicity and angiogenesis, promote cell migration and invasion, promote neurosphere formation and angiogenesis [11]. PTPRZ1-MET induces gliomas through elevated expression levels of MET mRNA, preserve fundamental properties of wild-type MET, and enhance phosphorylation [13].
PTPRZ1-NFAM1 fusion could be harbored in spitzoid melanoma[14]. This fusion was associated with elevated expression of the NFAT activated protein with ITAM motif 1 (NFAM1), and the protein encoded by NFAM1 contains an immune-receptor tyrosine-based activation motif that is thought to regulate the development of B cells[15].
Table 1
PTPRZ1-fusion genes
Figure 1
..| Band 1 | Band 2 | Fusion | Partner 1 | Partner 2 | cancer |
|---|---|---|---|---|---|
| 7q31 | 2q21 | ARHGEF4/PTPRZ1 | PTPRZ1 | ARHGEF4 | |
| 7q31 | 7q11 | CLIP2/PTPRZ1 | PTPRZ1 | CLIP2 | Brain Lower Grade Glioma |
| 7q31 | 3p21 | CSPG5/PTPRZ1 | PTPRZ1 | CSPG5 | Brain Lower Grade Glioma |
| 7q31 | 1q42 | GALNT2/PTPRZ1 | PTPRZ1 | GALNT2 | Skin Cutaneous Melanoma |
| 7q31 | Xp11 | HUWE1/PTPRZ1 | PTPRZ1 | HUWE1 | Brain Lower Grade Glioma |
| 7q31 | 7q22 | KMT2E/PTPRZ1 | PTPRZ1 | KMT2E | Stomach Adenocarcinoma |
| 7q22 | 7q31 | MLL5/PTPRZ1 | MLL5 | PTPRZ1 | Stomach Adenocarcinoma |
| 7q31 | 16q21 | PTPRZ1/ADGRG1 | PTPRZ1 | ADGRG1 | Brain Lower Grade Glioma |
| 7q31 | 1q22 | PTPRZ1/CCT3 | PTPRZ1 | CCT3 | Brain Lower Grade Glioma |
| 7q31 | 7q31 | PTPRZ1/DOCK4 | PTPRZ1 | DOCK4 | Malignant melanoma |
| 7q31 | 17p11 | PTPRZ1/EPN2 | PTPRZ1 | EPN2 | Brain Lower Grade Glioma |
| 6q21 | 7q31 | PTPRZ1/FYN | PTPRZ1 | FYN | Brain Lower Grade Glioma, Glioblastoma Multiforme |
| 7q31 | 17q21 | PTPRZ1/GFAP | PTPRZ1 | GFAP | Glioblastoma Multiforme |
| 7q31 | 1q25 | PTPRZ1/HMCN1 | PTPRZ1 | HMCN1 | |
| 7q31 | 7q31 | PTPRZ1/MDFIC | PTPRZ1 | MDFIC | Stomach Adenocarcinoma |
| 7q31 | 7q31 | PTPRZ1/MET | PTPRZ1 | MET | Glioblastoma Multiforme, Brain Astrocytoma, grade I-II |
| 7q31 | 22q13 | PTPRZ1/NFAM1 | PTPRZ1 | NFAM1 | Lung, Heart, Skin, Melanocytic neoplasm |
| 7q31 | 19p13 | PTPRZ1/PIN1 | PTPRZ1 | PIN1 | |
| 7q31 | 7q31 | PTPRZ1/PTPRZ1 | PTPRZ1 | PTPRZ1 | |
| 7q31 | 19p13 | PTPRZ1/SMARCA4 | PTPRZ1 | SMARCA4 | Brain Lower Grade Glioma |
| 7q31 | 7q32 | PTPRZ1/SND1 | PTPRZ1 | SND1 | |
| 7q31 | 7q11 | PTPRZ1/SRRM3 | PTPRZ1 | SRRM3 | Malignant melanoma |
| 7q31 | 7q32 | PTPRZ1/TSPAN33 | PTPRZ1 | TSPAN33 | Head and Neck Squamous Cell Carcinoma, Oral cavity Squamous cell carcinoma |
| 7q31 | 7q32 | PTPRZ1/UBE2H | PTPRZ1 | UBE2H | Breast Adenocarcinoma |
| 7q31 | 7q31 | ST7/PTPRZ1 | ST7 | PTPRZ1 | Prostate Adenocarcinoma |
| 7q31 | 19p12 | ZNF708/PTPRZ1 | PTPRZ1 | ZNF708 https://www.ncbi.nlm.nih.gov/protein/NP_067092.2 |
The structure and ligands of PTPRZ1
Several splice variants of PTPRZ1 have been discovered, including PTPRZ1-A (9.4 kb), a full-length transmembrane receptor form; PTPRZ1-B (6.4 kb), a short transmembrane receptor form with a deletion in the extracellular region; the other two are soluble secreted forms that lack both a transmembrane region and tyrosine phosphatase activity (phosphacan, short (4 kb) and long (8.4 kb; also known as phosphacan, PTPRZ-S)) [1, 2, 6, 16-18]. PTPRZ1-A and PTPRZ1-B are composed of several modular structural domains, including an N-terminal carbonic anhydrase-(CAH) like domain, a fibronectin type III domain, a serine, glycine-rich domain for chondroitin- sulfate (CS) attachment, a transmembrane segment, and two protein tyrosine phosphatase (PTP) domains plus C-terminal PDZ-binding motif [1, 16].The membrane proximal phosphatase domain (D1) is active, but the distal D2 domain is inactive[19]. A novel PTPRZ1 receptor sub-isoforms without a seven-amino acid sequence encoded by exon 16 was identified, which forms a part of the helix-turn-helix segment called the 'wedge' structure and is located at the N-terminal region in the membrane-proximal protein tyrosine phosphatase domain[20].
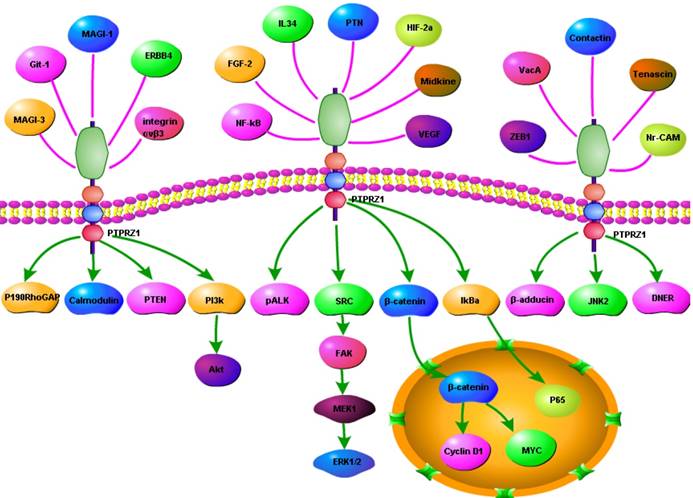
The extracellular region of PTPRZ1 binds to several cell molecules, including various secretory factors (pleiotrophin (PTN), midkine (MK), interleukin-34 (IL-34)), extracellular matrix proteins (tenascin- C and tenascin-R), and neuronal cell adhesion molecules (NrCAM, L1/Ng-CAM, F3/contactin, neural cell adhesion molecule, contactin-1 and contactin-2/ TAG-1/axonin-1) as well as amphoterin and fibroblast growth factor-2 (FGF-2). [3, 5, 6, 21-24].
PTN
PTN, IL-34 and midkine (MK) interact with the glycosaminoglycan structures of PTPRZ1, have been shown to function as inhibitory ligands against the tyrosine phoshatase activity of PTPRZ1 [21, 22]. PTN is the most potent known natural ligand of PTPRZ1 and strongly inhibits its activity putatively through ligand-induced dimerization of the active D1 domain of PTPRZ1[25]. The C-terminal tail of PTN is essential for maintaining stable interactions with chondroitin sulfate (CS) A, the type of CS commonly on PTPRZ1, which explained why truncated PTN missing the C-terminal tail is unable to signal through PTPRZ1 [26]. PTN-mediated PTPRZ1 oligomerization, which might inhibit PTPRZ1's phosphatase activity may involve both core protein and glycan components of PTPRZ1 [27]. PTPRZ1 signals through 'ligand-dependent receptor inactivation' of PTN and disrupts its normal role in the regulation of tyrosine phosphorylation of downstream signalling molecules [6, 28]. PTPRZ1-PTN interaction results in accumulation of tyrosine phosphorylation of multiple downstream proteins including SRC kinase, calmodulin, anaplastic lymphoma kinase (ALK), GIT1/Cat1, β-catenin, β-adducin, Fyn, GIT1/Cat-1, and P190RhoGAP, which in turn have been related to activation of multiple pro-tumorigenic signaling cascades[4, 6, 8, 28, 29]. PTPRZ1-PTN interaction modulates the tyrosine phosphorylation of factors that control cell-matrix interactions, cell-cell adhesion, cell migration, tumor cell invasion, tumor growth and metastasis [2, 4, 5, 29-32].PTPRZ1, syndecan-3 (N-syndecan, SDC3), and ALK have been characterized as PTN transmembrane receptors and to be implicated in its signaling [31-34]. PTN activates its cell surface receptors, regulating multiple functions including cell adhesion, cell migration, cell proliferation and cytoskeletal stability and negatively regulated PTPRZ1[35]. A more recent study isolated and characterized two forms of PTN, a full-length form (PTN18) which induced haptotactic migration, whereas the shorter (PTN15) activated a mitogenic signal transduction pathway. PTN15 stimulation through ALK activates a mitogenic signal transduction pathway, whereas binding of immobilized PTN18 to PTPRZ1 induces cell migration[32]. PTN has been shown to activate both the mitogen- activated protein kinase (MAPK) and the phosphatidylinositol 3-kinase (PI3K)-Akt signaling axes, and inhibitors of Erk1/2 or PI3K inhibit DNA synthesis stimulated by PTN [34].
IL-34
PTPRZ1, was an additional IL-34 receptor [3]. IL-34 is a 241 amino acid protein, which exists in two isoforms, differing by the addition of a glutamine inserted between position 80 and 81 in the 241 aa isoform, and generated by alternative splicing. IL-34 is formed by four α-helix and disulfdes bonds that lead to the formation of a homodimeric protein. IL-34 is a cytokine relatively conserved between species with 99.6% homology between human and chimpanzee. IL-34 selectively interacts with PTPRZ1 in membrane lysates through low affinity interactions with chondroitin sulphate chains, which suppresses their tumorigenic properties in a PTPRZ1 dependent manner[24]. Studies have identified roles for IL-34 in areas as remote as neuronal protection, bone-degenerative diseases, delayedtype hypersensitivity, infection, cancer, and more recently transplantation [36]. IL-34 binds specifically to PTPRZ1 to initiate downstream signaling, further inhibits cell proliferation, clonogenicity, and motility[24].Midkine (MK)
The C-terminally located C-domain of MK, a 13-kDa heparin-binding growth factor with 45% sequence identity to PTN, exhibited full activity for PTPRZ1 binding[37]. PTPRZ1 was involved in the haptotactic migration of UMR106 cells induced by midkine, which is highly related to PTN and binds to PTPRZ1 with high affinity [23]. MK enhances cell survival, cell migration, cytokine expression, cell differentiation, and involves in several physiological processes, including cancer, inflammation/immunity, blood pressure, development and tissue protection [38, 39].β-catenin
β-catenin is of major importance in cell-cell adhesion, cytoskeletal structure, and signaling pathways in normal and transformed cell growth[40]. The interaction of PTPRZ1 and PTN increases the steady-state tyrosine dephosphorylation of β-catenin, which interacts with the catalytically active D1 domain of PTPRZ1[25, 28, 40-42]. This process influences the cytosolic and nuclear availability of β-catenin, and activated the β-catenin pathway [42]. In the nucleus, β-catenin displaces the transcriptional repressor Groucho/TLE by binding to TCF3 (TCF7L1) and TCF4 (TCF7L2), thereby initiating transcription [25]. PTPRZ1 increases the expression of β-catenin in the nuclear fractions and further increases the activation by β-catenin of T cell factor (TCF)- dependent transcription [25]. Mcclain et al. found PTPRZ1 inhibition also potentiated GSK3β-regulated β-catenin translocation and two pools of β-catenin, which are regulated by PTPRZ1 and GSK3β are partially but incompletely overlapping, with GSK3β appearing to regulate β-catenin availability beyond that influenced by PTPRZ1 [25]. PTPRZ1 also regulates proliferation, tumorigenesis and E-cadherin-mediated cell-cell adhesion by activating or dephosphorylating β-catenin and its downstream genes, such as cyclin D1, c-myc, c-jun, fra-1, and CD44 [7, 35, 41, 42].MAGI, ERBB4 and PSD95
The interaction between PTPRZ1 and MAGI-1/ 3, PSD95 was found to be mediated primarily via the PDZ domain in vivo [23, 43]. The third PDZ domain of MAGI-1 is responsible for interaction with the carboxyl terminal PDZ-binding motif of PTPRZ1 [1]. Simultaneous binding of MAGI to PTPRZ1 and ERBB4 forms a complex. MAGIs can bring ERBB4 tyrosine kinase and PTPRZ1 tyrosine phosphatase to domains in the cell membrane, modulating the balance among signaling pathways downstream of these molecules [16]. An intracellular carboxyl-terminal region of PTPRZ1 pulled-down PSD95 and ERBB4 and the tyrosine phosphorylation level of ERBB4 was increased in PTPRZ1 gene deficient mice. Phosphorylation of ERBB4 was raised by PSD95, which was repressed by additional PTPRZ1 expression [44].Others
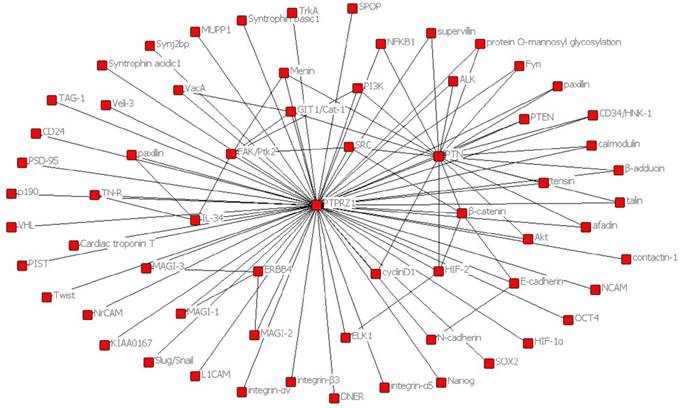
One HIF-2-specific gene was PTPRZ1 gene, which was upregulated about six-fold by HIF-2α[45]. HIF-1α and HIF-2α can bind to hypoxia-responsive elements 4 (HRE4) of the PTPRZ1 gene promoter, and the HIF-2-specificity of PTPRZ1 is mediated at least in part by cooperative binding of HIF-2 and ELK1 to nearby sites in the PTPRZ1 gene promoter [7]. PTPRZ1-D1902A and Git1 form a stable complex in mammalian cells, Git1 bound to PTPRZ1-D1902A dependent on the tyrosine-phosphorylated Git1, which was dephosphorylated by PTPRZ1 [23]. Endogenous VEGF formed complexes with PTPRZ1, which suggested PTPRZ1 was a cell membrane binding molecule for VEGF165 [46]. Chromatin immunoprecipitation assay identified the direct binding of NF-κB to the PTPRZ1 gene promoter, and clofibrate abolished this binding[47]. The CAH domain of PTPRZ1 has been shown to bind specifically to contactin [6]. Contactin is a 140 kDa glycosylphosphatidylinositol (GPI) membraneanchored neuronal cell recognition protein. Binding of glial PTPRZ1 to the contactin/Nr-CAM complex is important for neurite growth and neuronal differentiation[6, 48]. Rokavec et al. found PTPRZ1 gene promoter contain conserved putative ZEB1- binding sites and ZEB1 bound to PTPRZ1 gene promoters in MCF-10a-T cells [49]. PTPRZ1 directly interacts with integrin αvβ3 on the cell surface, and integrin αvβ3 interacted with both the long (380 kDa) and short (220 kDa) transmembrane isoforms of PTPRZ1, as well as with a transmembrane isoform of lower molecular mass (80-90 kDa). The longer PTPRZ1 isoforms interacted specifically with αv, while the 80-kDa band interacted specifically with the β3 subunit[50]. PTPRZ1 provides VacA-binding sites at least in part in the gastric epithelium and binding analysis indicated that VacA directly binds to PTPRZ1-B[51]. PTPRZ1 also binds to FGF-2, NCAMs, tenascin amphoterin and tenascin-C [5, 30]. And some other relevant molecules were identified using data from Cytoscape_3.6.0 (figure 3).The functions of PTPRZ1
Tumor cell proliferation and tumor growth
PTPRZ1 seems to be important for maximum tumor growth in situations of contact independence or in complex cellular environments, and blocking the PTPRZ1-PTN signaling potently suppressed glioblastoma growth and prolonged animal survival. PTN dephosphorylates the constitutively active PTPRZ1, which upon binding PTN dimerizes and becomes inactivated, resulting in an increase in tyrosine- phosphorylated β-catenin[28]. It further leads to increased levels of cytoplasmic β-catenin which translocates to the nucleus where it binds to the transcription factor TCF/LEF; this causes increased expression of MYC and Cyclin D1, stimulating cell cycling and proliferation [5]. Knock-down of PTPRZ1 decreased the amounts of nuclear β-catenin and suppressed cellular proliferation concomitant with a decrease in the expression of target genes: cyclin D1, c-myc, c-jun, fra-1, and CD44 in renal cell carcinoma [41]. However, studies in osteosarcoma were different from that in glioblastoma, and PTPRZ1-deficiency significantly enhanced osteosarcoma development with 19% Trp53-heterozygous mice, which suggest PTPRZ1 could function as a tumor suppressor function for osteosarcoma[21].
Figure 3
Relevant molecules that interacted with PTPRZ1 from Cytoscape_3.6.0 (figure 3).
 (Click on the image to enlarge.)
(Click on the image to enlarge.)
 (Click on the image to enlarge.)
(Click on the image to enlarge.)Cell-cell adhesion and cell migration
Cell-cell adhesion requires members of the cadherin-catenin families to link the highly conserved cadherin cytoplasmic domain to the actin-based cytoskeleton and to connect adjacent cells via the cadherin extracellular domains [28]. PTPRZ1-PTN interaction in liver cells modulates the tyrosine phosphorylation of factors, such as Src, focal adhesion kinase (FAK), PTEN, MEK, and Erk1/2, that control cell-cell adhesion, cell-matrix interactions and cell migration, including an increase in tyrosine- phosphorylated β-catenin, which is known to cause loss of cell-cell adhesion [28, 31, 52].PTPRZ1-PTN is involved in regulating cell migration and metastasis on various substrata, including FAK, PI3K, MAPK, Src, PTEN, MEK, and Erk1/2, and integrin αvβ3 activity [2, 17, 29, 31]. PTPRZ1 gene knockdown increased the ability of human prostate cancer cells to migrate and invade in vitro and to metastasize in vivo [31]. PTPRZ1 gene knockdown interrupts hydrogen peroxide-induced cell migration[53] and PTPRZ1 gene knockdown in human umbilical vein endothelial cells (HUVEC) inhibited PTN-induced migration and tube formation on matrigel[34]. Treating PTPRZ1-positive ductular progenitor cells with PTN caused time-dependent accumulation of many tyrosine-phosphorylated proteins, including talin, tensin, paxillin, afadin and supervillin. These latter proteins control cell migration, and PTPRZ1-PTN interactions directly regulate the migratory activity of cultured human stem cells and liver progenitor cell [29].
Epithelial-to-mesenchymal transition (EMT)
PTPRZ1 was a negative regulator of EMT, where cancer cells lose polarity, cell-cell contacts and other epithelial characteristics, switching to a motile mesenchymal phenotype. Common EMT features include a loss of E-cadherin, elevated N-cadherin, integrins, Twist, Slug, and Snail expression, increased nuclear β-catenin, and gain of fibroblastoid morphology [31]. The EMT is a critical event in early development and a critical component of progression of malignant cells to a more aggressive phenotype [54]. In prostate cancer, the loss of PTPRZ1 gene expression initiated EMT and increased the ability of the cells to migrate and invade[31]. Phalloidin staining of PTPRZ1 gene knockdown cells revealed increased stress fiber formation and a concomitant decrease in the number of cortical actin fibers, marker of lamellipodia formation and a shift in cadherin expression from the E to the N-cadherin form, known to characterize the EMT-like phenotype and to promote cell motility[31].Cancer stem cells
Tumor-associated macrophages (TAMs) secrete abundant PTN to stimulate glioma stem cells (GSCs) through PTPRZ1 thus promoting GBM malignant growth, and disrupting PTPRZ1 abrogated GSC maintenance and tumorigenic potential [55]. Knocking down PTPRZ1 gene altered the expression levels of SOX2, OLIG2, and POU3F2 and decreased the sphere- forming abilities in glioblastoma cells [19]. Another study found the knockdown of PTPRZ1 gene led to reduction of OCT4 and SOX2 transcripts by approximately 40%. OCT4 knockdown also resulted in 90% decrease of PTPRZ1 gene transcripts, consistent with the marked decrease of PTPRZ1 in differentiating embryonic stem cells[56]. PTN signaling via PTPRZ1 inhibits cell migration and acquisition of other mesenchymal cell characteristics, while PTPRZ1 alone promotes a migratory/mesenchymal phenotype when unopposed by PTN[52].Others
The expression of PTPRZ1 was lower in the clofibrate plus radiation group than in the irradiated group in pancreatic cancer cells[47]. PTPRZ1 was involved in radiosensitization and downregulated after PPARα activation by clofibrate to sensitize pancreatic cancer cells to radiation [47]. PTPRZ1-/- mice demonstrated marked social alteration with increased aggressivity, reminiscent of some positive symptoms of schizophrenia[57]. PTPRZ1-PTN also controls the timing of oligodendrocyte precursor cell differentiation, in which the CS moiety of PTPRZ1 maintains them in a monomeric active state until its ligand binding[22].The network of PTPRZ1
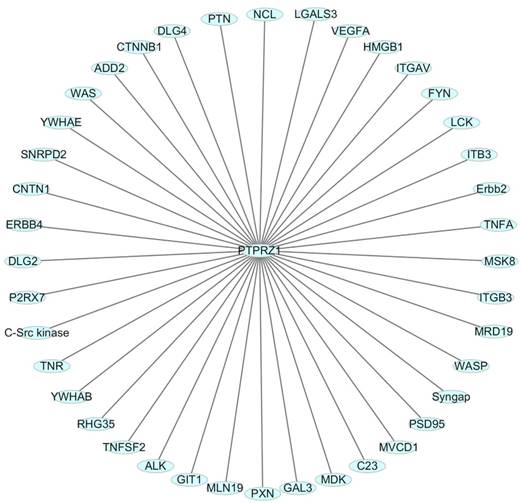
In order to present the interaction of PTPRZ1 with other molecules, the network of PTPRZ1 based on the literatures we reviewed we conducted (Figure 4, Figure 5). Ectopic menin expression significantly repressed PTN transcription, and indirectly inhibited PTPRZ1 expression through repressing PTN expression, and further inhibits FAK and ERK1/2 phosphorylation[35]. PTPRZ1 is also a substrate for protein O-mannosyl glycosylation by POMGnT1 [58]. Rokavec et al. found inhibition of miR-200c in MCF-10a cells decreased PTPRZ1 expression, and overexpression of PTPRZ1 in MCF-10a-T cells decreased phosphorylation and increased expression of IkBa, resulting in accumulation of p65 in the nucleus of MCF-10a-T cells[49]. Fukada et al. identified several other substrates: Git1, p190, PIST, MAGI-3, Veli-3, Synj2bp, Syntrophin acidic1, Syntrophin basic1, MUPP1, Cardiac troponin T, hDKFZp434G232, KIAA0167, SPOP by a genetic method named the ''yeast substrate-trapping system''[43]. Additional targets of the PTPRZ1-PTN signaling pathway thus far identified include calmodulin, Fyn(Src family Kinase), β-adducin, P190RhoGAP, Notch-related transmembrane protein Delta/Notch-like EGF-related receptor(DNER), PI3K, Akt, FAK, ERK1/2, paxillin, PTEN, PIST, integrin-α5, integrin-αv, and integrin-β3, PKCα/β and IkBa, to functions of different cellular systems[1, 2, 5, 8, 16, 22, 24, 25, 29, 31, 34, 43, 49, 54, 59].Survival
Based on TCGA data, PTPRZ1 gene might be associated with overall survival (OS) in bladder urothelial carcinoma (BLCA), BRCA, CESC, COAD, GBM, LGG, liver hepatocellular carcinoma (LIHC), LUAD, pancreatic adenocarcinoma (PAAD), and SKCM, PTPRZ1 gene might be associated with disease free survival (DFS) in BLCA, CESC, COAD, head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), LGG, LUSC, mesothelioma (MESO), PAAD, READ, SKCM, thymoma (THYM), uterine carcinosarcoma (UCS) and uveal melanoma(UVM). High PTPRZ1 gene expression might be associated with better OS in BRCA, CESC, LUAD, PAAD, and SKCM, and low PTPRZ1 expression might be associated with better OS in BLCA, COAD, GBM, LGG, LIHC. High level of PTPRZ1 gene was associated better DFS in CESC, HNSC, LUSC, MESO, PAAD, SKCM, THYM, and UCS, while low level of PTPRZ1 gene was associated better DFS in BLCA, COAD, KIRC, LGG, READ and UVM (Figure 6).PTPRZ1 was involved in survival signaling[60]. In human breast cancer, high expression of PTPRZ1 may be an independent risk indicator for triple- negative breast cancer (TNBC) recurrence and metastasis[61]. PTPRZ1 has a significant impact on survival of patients with oral squamous cell carcinoma, and patients with positive PTPRZ1 was associated with 8 times lower risk of death within 5 years than those with negative PTPRZ1[62].
The therapeutic values of PTPRZ1
PTPRZ1 might be a therapeutic target due to its role in cancer development, progression, and metastasis. Several inhibitors of PTPRZ1 or antibodies, including MY10, MY33-3[63], NAZ2329[19], were identified. Fujikawa et al. identified NAZ2329, a cell-permeable small molecule that allosterically inhibits PTPRZ1, which reduced the expression of SOX2 and abrogated the sphere-forming abilities in glioblastoma cells [19]. Protamine (PRM, a heparin antagonist) bound to the extracellular region of PTPRZ1, and functioned as a ligand mimetic of PTPRZ1, thereby suppressing its negative regulatory activity on oligodendrocyte differentiation[64]. However, few of them were studied in oncology field. Foehr et al. developed antibodies directed to PTPRZ1, which used the recombinant extracellular domain of human short PTPRZ1 to immunize mice and generate monoclonal antibodies that selectively recognize PTPRZ1 and bind to the antigen with low nanomolar affinities. These antibodies killed glioma cells in vitro when coupled to the cytotoxin saporin either directly or via a secondary antibody and significantly delay human glioma tumors in a mouse xenograft model[30]. Fujikawa et al. showed the intracellular delivery of SCB4380, the first potent inhibitor for PTPRZ1, by liposome carriers inhibited PTPRZ1 activity in glioblastoma cells, and thereby suppressed cell migration and proliferation in vitro and tumor growth in a rat allograft model[65]. Midkine and PTPRZ1 expression was detected in the gastric mucosal, submucosal and muscle layers, and rebamipide may protect the gastric mucosa by regulating midkine and PTPRZ1 expression[66].Discussion
PTPRZ1 is a transmembrane protein tyrosine phosphatase and expressed in different tumor tissues. Based on TCGA data, PTPRZ1 was up-regulated in CESC, GBM, LGG, LUSC, and down-regulated in BRCA, COAD, PRAD, READ and STAD. Given the strong up-regulation of PTPRZ1, antagonization of PTPRZ1 expression and/or signaling may be a promising strategy to inhibit the tumor growth. For other cancers in which PTPRZ1 is down-regulation, activating PTPRZ1-dependent signaling could specifically target tumor cells. These indicate that pharmacological inhibition or activation of PTPRZ1 is a promising strategy for the treatment of several tumors. There were several drugs that target PTPRZ1, including PTPRZ1 antibodies[30], inhibitors (MY10, MY33-3[63], NAZ2329[19], protamine[64], and SCB4380[65]) and activators (rebamipide[66]). Most of available studies focused on the glioma therapy, except rebamipide. Rebamipide was found to increase PTPRZ1 and MK in stomach tissues and cell lines, but its role in cancer was not certificated, especially in those cancers that PTPRZ1 was down-regulated. These studies were conducted in vitro, and no clinical trials or observational studies were found till now. There is still a long way to go to translate available research results in vitro into clinical practice.PTPRZ1 gene mutation accounts for a high proportion of cancer patients, especially in UCEC, the proportion could be up to 16.79%. Such a high proportion of cancer patients have PTPRZ1 mutations, so whether PTPRZ1 mutations could predict therapy efficacy or not deserved further research. Could the drugs that target PTPRZ1 be effective when PTPRZ1 was mutated? Or did PTPRZ1 mutation predict response of available treatment? Based on available literatures, not such a drug was found. Also no clinical or observational studies were found to assess the values of PTPRZ1 gene mutation in cancer therapy. In the future, the value of PTPRZ1 gene mutation in cancer therapy deserved to be researched and drugs that target patients with PTPRZ1 mutations were studied in literature. Tumors from patients harboring PTPRZ1-MET-fused glioblastoma are resistant to temozolomide therapy and have significantly compromised overall survival rates [11, 12]. Searching clinicaltrials.gov, only one trial about PTPRZ1 was retrieved without any study results, which was a phase I, open-label, dose-escalation study of c-Met inhibitor PLB1001 in patients with PTPRZ1-MET fusion gene positive recurrent high-grade gliomas. The prevalence, therapeutic and prognostic values of PTPRZ1-fusion genes in cancers were unclear and this needs further more researches.
The expression of PTPRZ1 in different kinds of cancer were different. This might imply different roles, however, the role of PTPRZ1 in cancer development, progression, and metastasis was not well studied. Based on available studies, PTPRZ1 functions in cell proliferation, cell adhesion and migration, EMT, cancer stem cells and treatment resistance by interacting with or binding to several molecules. The functional significance of the binding of molecules in PTPRZ1 signaling and their molecular pathways still remains unclear[2, 22]. Based on our review, these included PTN, midkine, interleukin-34, β-catenin, VEGF, NF-κB, HIF-2, PSD-95, MAGI-3, contactin and ErbB4. One of the most important ligands was PTN, and the PTPRZ1-PTN network was complex, which could activate PI3K and MAPK signaling pathway. Although Fukada et al. identified several other substrates by a genetic method named the ''yeast substrate-trapping system''[43], few of them were well studied in available literatures. More researches are needed to study what and how PTPRZ1 interacts with.
Based on TCGA data, PTPRZ1 gene might be associated with OS and DFS in several kinds of cancer, which implies that PTPRZ1 was involved in survival signaling[60]. The prognostic values of PTPRZ1 in different kinds of cancer were inconsistent, which imply that PTPRZ1 might play different roles in cancer progression. Our previous study showed high expression of PTPRZ1 may be an independent risk indicator for triple negative breast cancer recurrence and metastasis[61]. PTPRZ1 has a significant impact on survival in patients with oral squamous cell carcinoma, and patients with positive PTPRZ1 was associated with 8 times lower risk of death within 5 years than those with negative PTPRZ1[62]. The prognostic values of PTPRZ1 expression in other kinds of cancer were not assessed in available literatures, and further studies were required.
Although there were lots of studies focusing on PTPRZ1 in oncology field, lots of aspects of PTPRZ1 were not well studied. First, the prevalence of PTPRZ1-fusion genes and PTPRZ1 protein in cancers were unclear and their values in drug therapy and prognosis needs further research. Future studies should screen drugs for patients with PTPRZ1 gene mutations, PTPRZ1 fusion genes, and PTPRZ1 protein positivity. Second, pharmacological inhibition or activation of PTPRZ1 is a promising strategy for the treatment of several tumors. However, most of available studies focused on inhibitors or antibodies that target PTPRZ1 about gliomas in vitro and whether they were effective in other kinds of cancer which have up-regulation of PTPRZ1 expression. None of them were translated into clinical practice and the most effective drugs were not clear. For those kinds of cancer which have down-regulation of PTPRZ1 expression, PTPRZ1 activation drugs were not tested in cancer. Third, due to different expressions of PTPRZ1 in different tumor tissues, so whether the PTPRZ1 could be a biomarker for diagnosing cancer and distinguishing different kinds of cancer or not was unclear. Although a recent study showed PTPRZ1 was significantly over-expressed at the mRNA and protein levels in grade II/III glioma patients as compared to non-tumor samples[67], no studies showed PTPRZ1 could be used as a diagnostic biomarker. So the role of PTPRZ1 in diagnosing cancer should be considered in the future.
Our review is a descriptive study which described the research results of PTPRZ1 comprehensively based on Pubmed and EMBASE. Also the expression of PTPRZ1 gene, gene mutations, gene fusions, and their prognostic values in different kinds of cancers were analyzed. In this review, we could find out PTPRZ1 is expressed in different tumor tissues, and functions in cell proliferation, cell adhesion and migration, EMT, cancer stem cells and treatment resistance by interacting with or binding to several molecules. These included PTN, MK, IL-34, β-catenin, VEGF, NF-κB, HIF-2, PSD-95, MAGI-3, contactin and ErbB4. The PTPRZ1 network was complex, which could activate PI3K and MAPK signaling pathway. PTPRZ1 was involved in survival signalling and could predict the prognosis of several tumors. PTPRZ1 might be a therapeutic target in several kinds of cancers, however, available studies focused on the PTPRZ1 target for GBM in vitro. There is still a long way to go to translate available basic research results into clinical practice.
Figure 6 Survival plot based on TCGA data (the plot